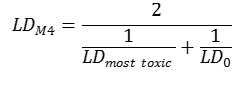similarity: the pair-wise distance between a query compound and three nearest neighbors in the training set is calculated by Pearson’s correlation coefficient in the space of the independent variables obtained after SCR (in AD if Pearson’s correlation coefficient < 0.7);
leverage (in AD if leverage for a compound < 99th percentile in the distribution of the leverage values calculated for the training set).
assessment of accuracy: the error of prediction for the three most similar compounds in the training set relative to the whole training set (in AD if RMSE3NN/RMSEtrain < 1).
Based on the metabolites acute toxicity values calculated by our method , we proposed four different methods for calculating the integrated toxicity.
Method 1. The integral toxicity is calculated as the effect of toxicity of all metabolites without taking into account toxicity of the parent compound. It is assumed that each of the parent compounds is metabolized and all its metabolites are formed with an equal probability Pi= 1/N (N is an amount of metabolites). We estimate an integral LD50 value LDM1 using the predicted LD50 values for metabolites (LDi) as follows:
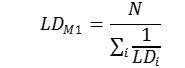
Method 2. The integral toxicity is calculated as the toxic effects of the parent compounds and all its metabolites. One assumes that the parent compound is not completely metabolized and would be considered together with its metabolites. We estimate an integral LD50 value LDM2 using the predicted LD50 values for metabolites (LDi) and for the parent compound (LD0) as follows:
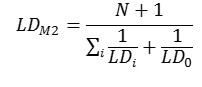
Method 3. The integral toxicity is calculated as the toxic effects of the most toxic metabolite. One assumes that effect of the most toxic metabolite is crucial, even if it is formed in a small quantity. We estimate an integral LD50 value LDM3 as follows:
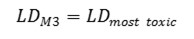
Method 4. The integral toxicity is calculated as the toxic effects of the most toxic metabolite and the parent compound. One assumes that both effects of the most toxic metabolite and the parent compound is important. We estimate an integral LD50 value LDM4 as follows: