
According to the World Health Organization (WHO) estimates, in 2017 about 36.9 million adults and children live with HIV/AIDS worldwide.
Application of Highly Active Antiretroviral Therapy (HAART) maximally suppresses the HIV virus, terminates the progression of HIV disease and prevents onward transmission of HIV.
However, the existing medicines do not provide the complete curation, exhibit serious side effects, and cause the appearance of resistant strains.
Therefore, continuous studies directed to the discovery of new more safe and potent anti-HIV agents are performed.
Application of the in silico predictions significantly reduces the time and financial expenses for finding new medicines.
Based on our 30+ year experience in development and application of ligand-based computational tools for wide range of (Q)SAR studies, we created the web-service AntiHIV-Pred.
It allows estimating the potential of the particular drug-like compound as a pharmaceutical agent for treatment of HIV-1 infection and its comorbidities.
The user has an immediate access to obtain predictions for compounds from the Open NCI database developed in the framework of the NCI Development Therapeutics Program, where extramural researchers may obtain samples for testing as potential human medicines.
Prediction of affinity to different HIV-1 targets is based on the quantitative structure-activity analysis of molecules contained in ChEMBL (version 24), Integrity and in NIAID databases.
Raw data were carefully curated according to the nowadays recommendations: the recognized duplicates were removed, data validity was verified using the information presented in the original publications, etc.
QSAR models were built using our computer program GUSAR, which good performance has been confirmed by the comparative computational experiments.
Reasonable accuracy and predictivity (view List of Predicted Activities) was achieved for estimation of both compound’s action on HIV-1 targets with GUSAR models and biological activities relevant to the treatment of HIV-associated comorbidities with computer program PASS.
Using as the input information structural formula of compound prepared with the ChemAxon Marvin JS chemical editor, one may obtain the results of prediction for molecule under study in one click.
Output results of predictions are provided as the estimated IC50 values for interaction of the molecule with HIV-1 protease, reverse transcriptase and integrase, and list of probable biological activities useful for the treatment of HIV-associated comorbidities with two probabilities Pa (estimate of belonging to the class of “actives”) and Pi (estimate of belonging to the class of “inactives”).
Predict biological activities
How to:
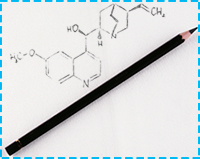
Draw your molecule
You may use the Marvin JS editor for drawing your molecule. Marvin JS runs in any HTML5-capable browser without any plugin. For IE, version 9 or above is needed for Marvin JS due to use of HTML5.
Predict HIV inhibiting activity
Select HIV molecular target and the source of experimental data used for model development. Click "Predict" button to predict whether a compound inhibits HIV molecular targets and has biological activities for HIV-associated comorbidities.
Click molecular editor and Draw your molecule

Select the appropriate HIV target
Select the model data source
PREDICTION EXAMPLES
Here we perform a prediction for structures in Open NCI database. Corresponding compounds could be ordered.
"One of the first approaches in the field of in silico pharmacology was PASS (Prediction of Activity Spectra for Substances), which applies a set of 2D descriptors to compounds that are then correlated with a set of bioactivities. "
Vegner L. et al. J. Med. Chem., 2013, 56: 8377. DOI: 10.1021/jm400813y
Try It Today
